Methylation Mark Atlas
An atlas of methylation marks identified by Specific Methylation Analysis and Report Tool (SMART) based on 50 methylomes across human cell lines and tissues
An atlas of methylation marks identified by Specific Methylation Analysis and Report Tool (SMART) based on 50 methylomes across human cell lines and tissues

| Search | Browser | Download | Analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MethySegments | Super-MethyMarks | Visual analysis of MethMarks via MethyMark Browser | Download pre-defined MethyMarks and association tracks | Use SMART to explore MethyMarks for your methylomes | |||||
| MethyMarks | Large-MethyMarks | ||||||||
 |
 |
||||||||
 |
|||||||||
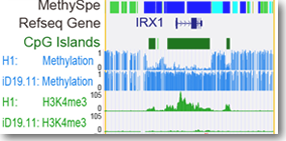
It has been demonstrated that cell-type-specific methylation distinguishes human cell-types and associates with cell-type-specific functions. This feature indicates the segments with high methylation specificity may be potential methylation marks of human cell-types. The collection of human methylomes enables the dissection of methylation marks across cell-types and tissues comprehensively sampled across the human body. The distance-dependent methylation similarity between neighboring CpGs which would be useful for identifying the genome regions consist of uniformly methylated CpGs or cell-specific methylated CpGs.
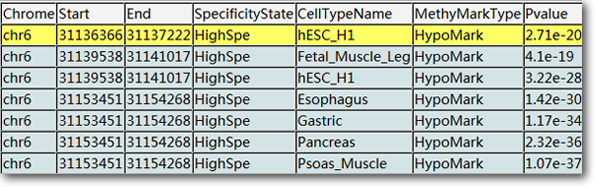
Thus, we developed entropy-based method to carry out methylome segmentation and identify the segments with high/low/no specificity across human cell-types. Then a statistical method were developed to identify the High/LowSpe segments with specific hypo-/hyper-methylation in the minority of cell-types as cell-type-specific hypo-methylation marks (HypoMarks) or cell-type-specific hyper-methylation marks (HyperMarks) . Using this method, we presented an atlas of cell-type-specific methylation marks (MethyMarks) across 42 cell-types of human, which are the union of 460, 438 MethyMarks across all these cell-types. MethyMarks overlapped with super-enhancers are treated as Super-MethyMarks and those with length >=3.5kb are Large-MethyMarks.All these information can be searched, viewed and downloaded for further analysis.
MethyMark seletor: Using the search engine, users can found the methymarks in the given genome locations or specific genes of intresting. For instance, seting the Chrome=chr6, Start=31124000, and End=31147000 in the page SuperMethyMarks results in the methymarks overlapped with super-enhancer. Using the similar method, the methylation segments by SMART, cell-type-specific methymakrs and those large methymarks can be obtained in the specific page.
MethyMark files dataload: In the download page, all data files including methylation segments, cell-type-specific MethyMarks, Super-MethyMarks and Large-MethyMarks are provided for local analysis, visualization in UCSC genome browser, and even building of functional databases related with these MethyMarks.
SMART software: According to process of identifying methylation marks across human cell-types in this paper, we presented a Specific Methylation Analysis and Report Tool (SMART) for BS-Seq data. SMART should be usefule for researchers in automated analyzing their own methylation data and identifing cell-types-specific or sample-specific methylation marks.
If you use this resource, please cite the paper
Hongbo Liu et al. Systematic identification and annotation of human methylation marks based on bisulfite sequencing methylomes reveals distinct roles of cell type-specific hypomethylation in the regulation of cell identity genes Nucleic Acids Res: 2016 ,44(1) ,75-94.